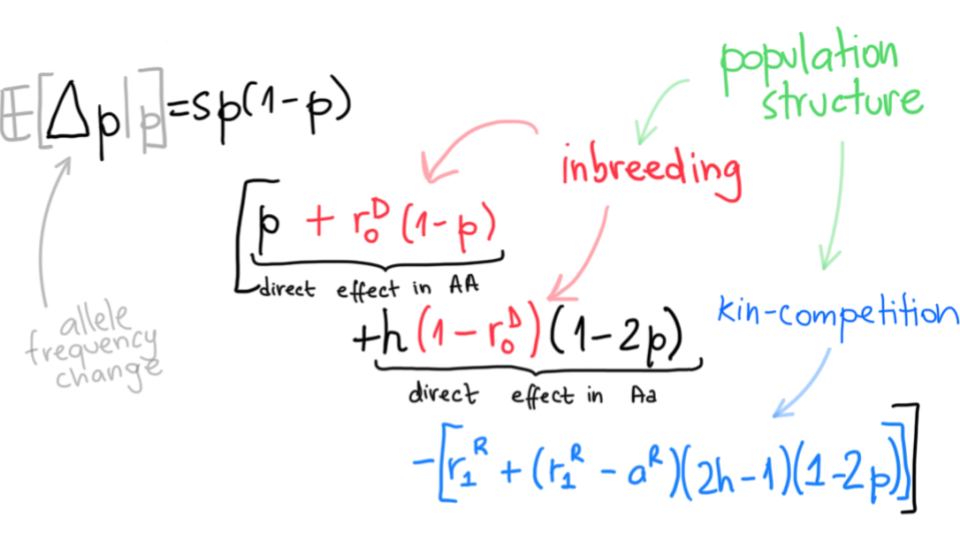
Due to non-random interactions such as inbreeding or local competition, most natural populations show some level of structure. We use mathematical models and computer simulations to investigate how such structure influences genetic evolution and variation within and between individuals. We have shown for instance how limited dispersal can accelerate the fixation of beneficial allele and thus influence the effects of such sweeps on variation at nearby loci. This happens where limited dispersal moderately increases effective population size (which slows fixation) but sufficiently increases homozygosity due to inbreeding, exposing rare recessive alleles to selection (which accelerates fixation). In addition, we show that limited dispersal tends to blur the genetic signatures of sweeps of non-additive alleles.
Population genetics of structured populations
Vitor Sudbrack
Relevant publications or preprints
-
Fixation times of de novo and standing beneficial variants in subdivided populations
Vitor Sudbrack and Charles Mullon Genetics, 2024.